5 Essential Tips for Mastering Restriction Enzyme Worksheets
Restriction enzymes, often referred to as molecular scissors, play an indispensable role in molecular biology and genetic engineering. They are proteins that cleave DNA at or near specific recognition nucleotide sequences known as restriction sites. For those diving into the intricacies of molecular biology, understanding and mastering the use of restriction enzymes can open up a world of genetic exploration and manipulation. Here are five essential tips to help you excel when working with restriction enzyme worksheets.
1. Understanding Restriction Enzyme Basics

Before diving into the practical aspects of restriction enzymes, a solid foundational knowledge is crucial:
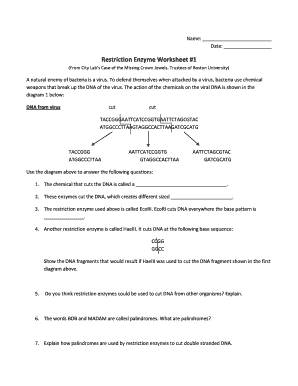
- Recognition Sequences: Each enzyme recognizes a specific DNA sequence, typically 4 to 8 base pairs long, known as its recognition site. For instance, EcoRI recognizes and cleaves at the sequence 5'-GAATTC-3'.
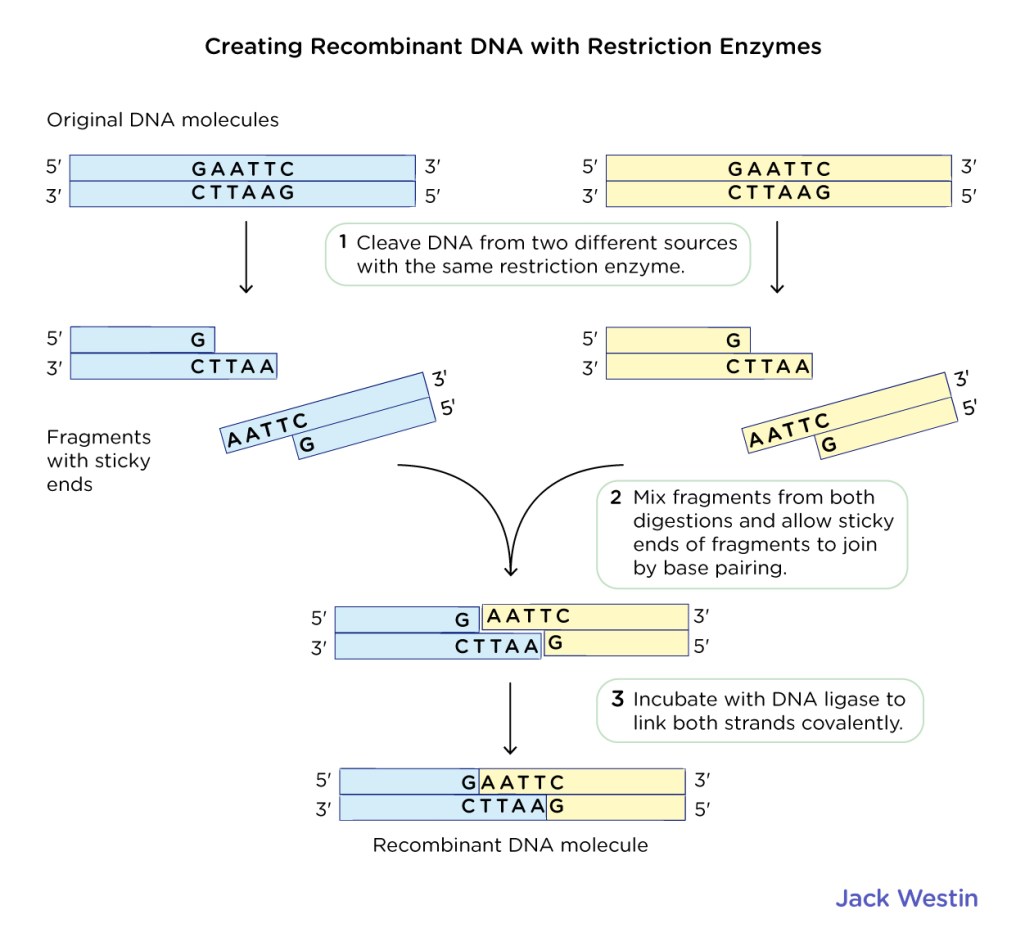
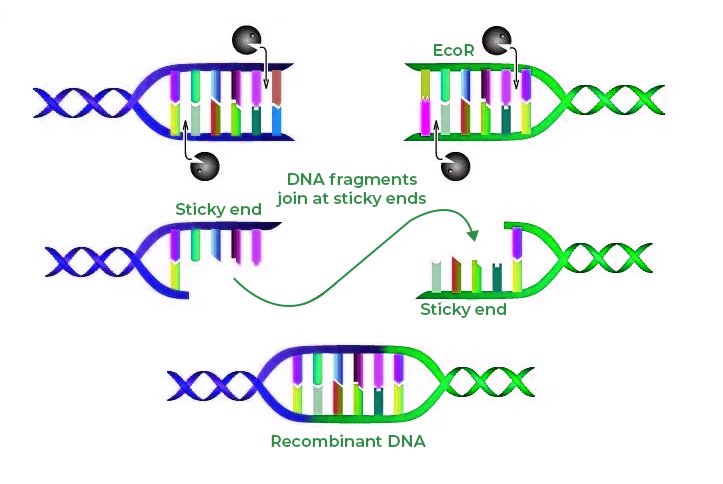
- Types of Cuts: Enzymes can make two types of cuts - blunt ends or sticky ends. Blunt end cutters produce ends without overhangs, whereas sticky end cutters leave small overhangs. Understanding this can affect downstream applications like cloning.
- Star Activity: Some enzymes can exhibit star activity under suboptimal conditions, where they cut at non-specific sites. Be aware of this potential, especially when conditions are not strictly controlled.
2. Selecting the Right Enzyme
Choosing the appropriate enzyme is paramount for efficient cutting:
- Sequence Analysis: Use software tools to analyze DNA sequences for restriction sites. Programs like NEBcutter or SnapGene can provide insights into enzyme suitability.
- Consider Overhang: If you're cloning, consider whether you need sticky ends or blunt ends, and choose enzymes that provide the necessary overhang for your ligation strategy.
- Buffer Compatibility: Enzymes often work best in specific buffers. Ensure that the chosen enzymes are compatible in terms of buffer conditions to streamline your experimental setup.
🔍 Note: Always check for isoschizomers - enzymes that cut at the same site but differ in buffer requirements or activity under various conditions.
3. Optimizing Reaction Conditions
Mastering the physical parameters of restriction enzyme digestion is key:
- Temperature: Most enzymes operate optimally at 37°C. However, some like NotI require 50°C, while others might work better at a lower temperature.
- Buffer and Salt: Buffers can affect the efficiency and specificity of the enzyme. Ensure your reaction mix is within the manufacturer's recommended salt concentration.
- Incubation Time: While most enzymes require 1 to 2 hours, ensure you incubate long enough to maximize digestion but not too long to risk star activity.
4. Post-Digestion Analysis

Once your digestion is complete, confirming the result is crucial:
- Agarose Gel Electrophoresis: This is the gold standard for checking if your digestion was successful. Run your DNA alongside appropriate DNA ladders to determine fragment sizes.
- Enzyme Inactivation: Some enzymes can be inactivated by heat (e.g., 80°C for 20 minutes for EcoRI). Ensure this step is done before running the gel to prevent false positives or negatives due to ongoing enzyme activity.
- Quantitative PCR or qPCR: For a more precise analysis, especially when dealing with multiple fragments, qPCR can help quantify DNA fragments post-digestion.
5. Trouble-Shooting Common Issues

Even with meticulous planning, issues can arise. Here are ways to troubleshoot common problems:
- Incomplete Digestion: Ensure sufficient enzyme activity, check buffer compatibility, and optimize DNA concentration. Sometimes, re-evaluating the enzyme units might be necessary.
- Star Activity: If you observe digestion at non-specific sites, consider if buffer conditions are optimal, enzyme concentration is correct, or if you've over-incubated the reaction.
- No Digestion: Review enzyme activity, check for inhibitors in your DNA preparation, and ensure DNA is not degraded or too concentrated, which can inhibit enzyme function.
The mastery of restriction enzyme techniques is fundamental for anyone interested in molecular biology, cloning, or genetic engineering. These tips provide a structured approach to dealing with restriction enzyme worksheets, which are vital for hands-on experience and understanding. By focusing on the basics, selecting the right enzymes, optimizing conditions, analyzing results, and troubleshooting, you can ensure your experiments yield the desired results, thereby enriching your molecular biology toolkit.
Remember, the journey through restriction enzymes is one of both patience and precision. With these tips, you're well on your way to becoming proficient in using these molecular tools to sculpt DNA as needed.
What is the importance of recognition sequences?
+
Recognition sequences are critical because they determine where the enzyme will cut the DNA. Each enzyme has a unique sequence, allowing for selective DNA manipulation, which is essential for processes like cloning, gene editing, and restriction mapping.
Can I use multiple enzymes in a single reaction?

+
Yes, double or multiple digests can be done if the enzymes have compatible buffer requirements. Ensure the enzymes are active in the same buffer and salt concentration, and the digestion conditions do not lead to star activity or inhibit one enzyme.
Why does temperature matter in enzyme digestion?

+
Temperature affects the enzyme’s efficiency and specificity. Most enzymes have an optimal temperature at which they work best, often around 37°C. Deviations can result in suboptimal cutting or star activity, where the enzyme cuts at non-specific sites.