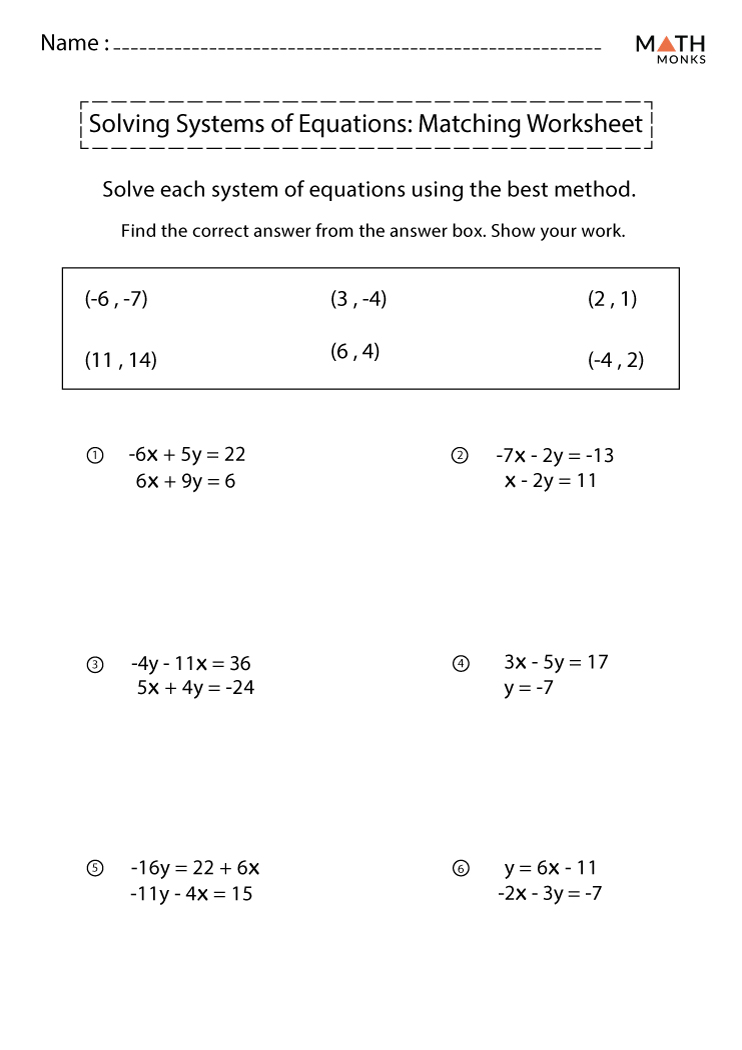
Restriction Enzyme Worksheet #1: Complete Answer Key Revealed

In the expansive world of molecular biology, understanding the function and application of restriction enzymes is fundamental. These enzymes, also known as restriction endonucleases, play a crucial role in DNA manipulation techniques like cloning, gene editing, and genetic engineering. This blog post will serve as an in-depth guide to understanding restriction enzymes through a comprehensive worksheet, offering insights into their use, functionality, and the key points to consider when working with them. Whether you're a student, researcher, or enthusiast, this post aims to equip you with the knowledge to tackle complex genetic problems with confidence.
What are Restriction Enzymes?

Restriction enzymes are proteins produced by bacteria that cleave DNA at specific recognition sites. Here's what you need to know:
- They act as molecular scissors, cutting DNA at or near specific nucleotide sequences known as restriction sites.
- These enzymes help bacteria protect themselves from viral infection by degrading the viral DNA.
- The most common type, Type II restriction enzymes, cut DNA precisely at their recognition sites, making them invaluable in biotechnology.
Restriction Enzyme Worksheet #1

This worksheet challenges students to predict the results of enzymatic digestion on various DNA sequences. Here, we'll provide answers and explanations:
Question 1:

Given the sequence of a plasmid with recognition sites for EcoRI, HindIII, and BamHI, predict the fragments that will result from digestion with each enzyme.
| Enzyme | Recognition Sequence | Fragments Produced |
|---|---|---|
| EcoRI | GAATTC | Two fragments: one from the start to the first site, and one from the first site to the end. |
| HindIII | AAGCTT | Fragments: From start to HindIII site, from HindIII site to BamHI, and from BamHI to the end. |
| BamHI | GGATCC | Fragments: From start to BamHI, and from BamHI to EcoRI. |

Question 2:

How does partial digestion differ from complete digestion, and how might this impact cloning strategies?
- Partial Digestion: Results in some of the recognition sites being cut, leaving longer fragments. This can be beneficial for creating overlap regions in vectors for gene insertion or ligation.
- Complete Digestion: All recognition sites are cut, producing smaller, more uniform fragments, which is ideal for mapping or analyzing a DNA sequence.
Key Considerations When Using Restriction Enzymes

When using restriction enzymes, several key factors influence their efficacy:
- Reaction Conditions: Optimal temperature, pH, and salt concentration must be maintained for specific enzymes.
- Enzyme Amount: Using the right enzyme concentration ensures complete digestion without nonspecific cleavage.
- Buffer Systems: Enzymes often require specific buffers to function correctly.
- Reaction Time: Sufficient time is necessary for complete digestion.
🔍 Note: Over-digestion might lead to star activity, where the enzyme cuts at unintended sequences.
Applications of Restriction Enzymes

Restriction enzymes are not just tools for cutting DNA; they are key in:
- DNA Cloning: Creating compatible ends for inserting genes into vectors.
- Restriction Fragment Length Polymorphism (RFLP): Identifying mutations or polymorphisms in DNA.
- DNA Mapping: Physical mapping of genomes by creating sequence landmarks.
- Forensic DNA Fingerprinting: Used in crime scene investigations to match DNA samples.
Understanding Enzyme Recognition Sites

Recognition sites for restriction enzymes are:
- Short, usually 4-8 base pairs long.
- Palindromic, meaning the sequence reads the same backward as forward on the complementary strand.
The specificity of these sites allows scientists to precisely edit or analyze DNA sequences. Here's an example:
EcoRI recognition site:
5' - G A A T T C - 3'
3' - C T T A A G - 5'
Advanced Techniques Involving Restriction Enzymes

Beyond basic cutting, restriction enzymes are involved in:
- Double Digestion: Using two enzymes simultaneously or sequentially to cut DNA at two different sites.
- Isoschizomers: Enzymes from different bacteria that cut the same recognition sequence.
- Neoschizomers: Enzymes that cut within a common recognition site but at a different location.
Final Thoughts

As we've explored, restriction enzymes are not just tools but are pivotal in shaping genetic research. Their precision and utility have led to advancements in gene therapy, agriculture, forensic science, and beyond. By understanding the mechanics, conditions, and applications of these molecular scissors, researchers can harness their power for innovative solutions in biotechnology. The study of restriction enzymes continues to evolve, offering ever-more sophisticated techniques for genetic manipulation and analysis. Remember, mastering these enzymes requires both knowledge and practice, making every experiment an opportunity to refine your skills in the delicate art of molecular biology.
What happens if I digest a plasmid with two different enzymes?

+
Digesting a plasmid with two enzymes results in fragments with different ends, which can reduce the likelihood of plasmid recircularization, making it useful for cloning purposes.
How do I choose which restriction enzyme to use?

+
Selection depends on the desired cut site, the frequency of the recognition sequence in your DNA of interest, and the compatibility of the overhangs for cloning.
Can restriction enzymes be used for genome editing?

+
While not primarily used for genome editing, enzymes like Cas9 in CRISPR systems function similarly to restriction enzymes but with programmable specificity, allowing for targeted genome edits.
What’s the significance of using isoschizomers?

+
Isoschizomers provide an alternative source for the same restriction site, useful if one enzyme becomes unavailable or shows different activity in your experimental conditions.
How do I prevent nonspecific DNA cleavage?

+
Use optimal reaction conditions, fresh enzyme stocks, and avoid long incubation periods or high enzyme concentrations to minimize the risk of star activity.