5 Ways to Understand Protein Folding and Structure

Protein folding and structure are fundamental topics in biochemistry and molecular biology. The way proteins fold and arrange themselves influences their function, interactions with other molecules, and their stability. Understanding these processes can provide insights into various diseases, biological mechanisms, and drug design. In this comprehensive guide, we'll explore five key approaches to understanding protein folding and structure:
1. Computational Modeling


Computational modeling involves using algorithms and software tools to predict how a protein will fold into its tertiary structure from its primary amino acid sequence. Here are some key aspects:
- Ab initio methods: These approaches predict folding without prior knowledge of protein structures, relying on physical forces like hydrogen bonding and hydrophobic effects.
- Homology modeling: If a protein shares significant sequence similarity with a protein whose structure is known, its structure can be modeled using the known structure as a template.
- Monte Carlo and Molecular Dynamics simulations: These techniques simulate the dynamic behavior of proteins over time, considering different conformations and their energy states.
💡 Note: Computational methods often require high computational power, especially for accurate predictions on larger proteins or complex assemblies.
2. X-ray Crystallography

X-ray crystallography is one of the most established methods for determining the atomic and molecular structure of a protein:
- Crystal Growth: Proteins are crystallized to form regular three-dimensional arrays.
- X-ray Diffraction: X-rays are diffracted by the crystal, producing a diffraction pattern.
- Data Interpretation: The diffraction pattern is analyzed to reconstruct the electron density of the protein, which is then used to deduce the molecular structure.
This method provides detailed, high-resolution images of protein structures, which are invaluable for understanding interactions, binding sites, and drug design.
3. Nuclear Magnetic Resonance (NMR) Spectroscopy


NMR spectroscopy allows for the study of proteins in solution, offering insights into:
- Dynamic Information: Unlike crystallography, NMR can observe the flexibility and movements of proteins.
- Protein-protein and Protein-ligand interactions: These can be studied through chemical shift mapping or NOE (Nuclear Overhauser Effect) spectroscopy.
- Non-crystalline Proteins: NMR can characterize proteins that do not easily crystallize.
NMR studies are complex and time-consuming but provide unique insights into protein behavior in a near-native environment.
4. Cryo-Electron Microscopy (Cryo-EM)


Cryo-EM has revolutionized structural biology by:
- Preserving Protein Structures: Proteins are flash-frozen, maintaining their native structures for imaging.
- Single Particle Analysis: This technique analyzes thousands of particles to build a 3D model without the need for crystals.
- Resolutions: Cryo-EM can achieve near-atomic resolution, which was not possible a few years ago.
Cryo-EM is particularly useful for large macromolecular assemblies and has become an indispensable tool in structural biology due to its ability to study dynamic processes.
5. Bioinformatics and Sequence Analysis


Bioinformatics uses computational tools to analyze protein sequences to:
- Predict Functional Sites: Sequence motifs and patterns often indicate functional domains or active sites.
- Assess Evolutionary Relationships: Comparative sequence analysis can reveal conservation patterns, suggesting structural or functional importance.
- Model Protein Interactions: Tools like protein-protein interaction networks help understand the context in which proteins operate.
Bioinformatics complements experimental methods by providing a vast array of data for analysis, enabling researchers to hypothesize and design targeted experiments.
The journey to understand protein folding and structure involves a combination of experimental techniques and computational analysis. Each method brings its strengths and limitations, making an integrated approach optimal for a comprehensive understanding. By employing these diverse strategies, researchers can unravel the intricate details of how proteins fold, interact, and function, providing insights into fundamental biological processes and aiding in the development of therapeutic interventions.
Why is protein folding important?
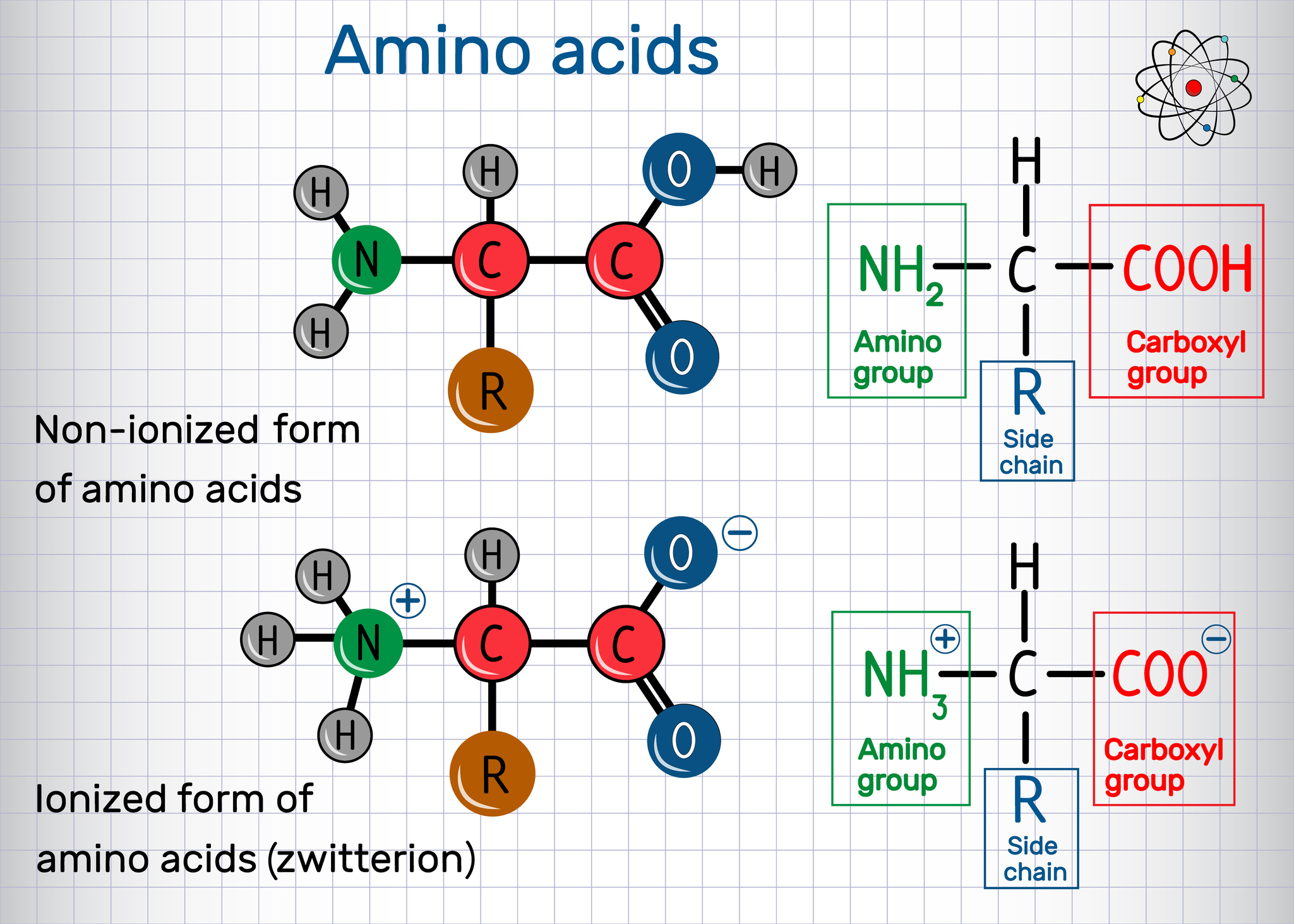
+
Protein folding is crucial because the three-dimensional structure of a protein determines its function. Improper folding can lead to diseases like Alzheimer’s or cystic fibrosis. Understanding folding helps in designing drugs that target specific protein structures or conformations.
How accurate are computational predictions of protein structure?
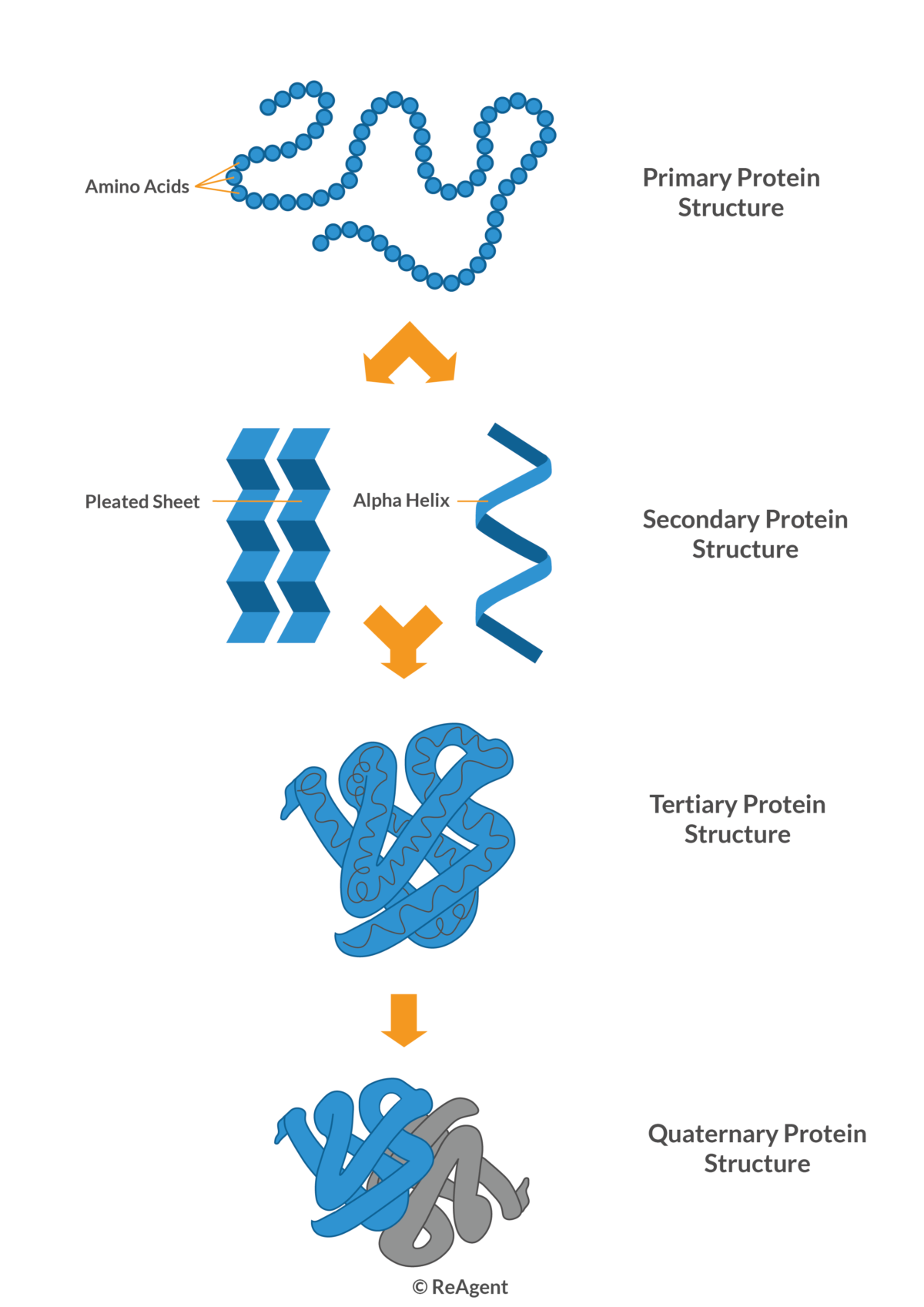
+
The accuracy of computational predictions depends on the method used and the available data. Tools like AlphaFold have shown remarkable accuracy when structural templates exist or when proteins have known homologs. For novel proteins without templates, accuracy can vary significantly.
What are the main differences between X-ray crystallography and NMR spectroscopy?

+
X-ray crystallography provides high-resolution static structures, requiring protein crystals, whereas NMR can observe proteins in solution, providing dynamic structural information but often at lower resolution. NMR doesn’t require crystallization, making it suitable for a broader range of proteins.



