Phylogenetic Tree of Trees Worksheet: Answer Key Revealed

In the expansive universe of biology, the tree of life is more than just a metaphor—it's a tangible representation of how species have evolved and diverged over millions of years. One of the fundamental tools in understanding this evolutionary history is the phylogenetic tree. This blog post aims to guide you through constructing and interpreting a phylogenetic tree, focusing on trees (the kind with leaves), and includes an answer key to a worksheet that reinforces these concepts.
What is a Phylogenetic Tree?

A phylogenetic tree, or evolutionary tree, depicts the relationships among different biological species based on their evolutionary relatedness and genetic characteristics. Here’s what it encompasses:
- Nodes: Represent speciation events or divergence points where a common ancestor gives rise to two or more descendant lineages.
- Branches: Symbolize the passage of time and evolution, with each branch representing a lineage or taxon.
- Tips: Represent current species or the most recent taxa included in the tree.
Constructing a Phylogenetic Tree
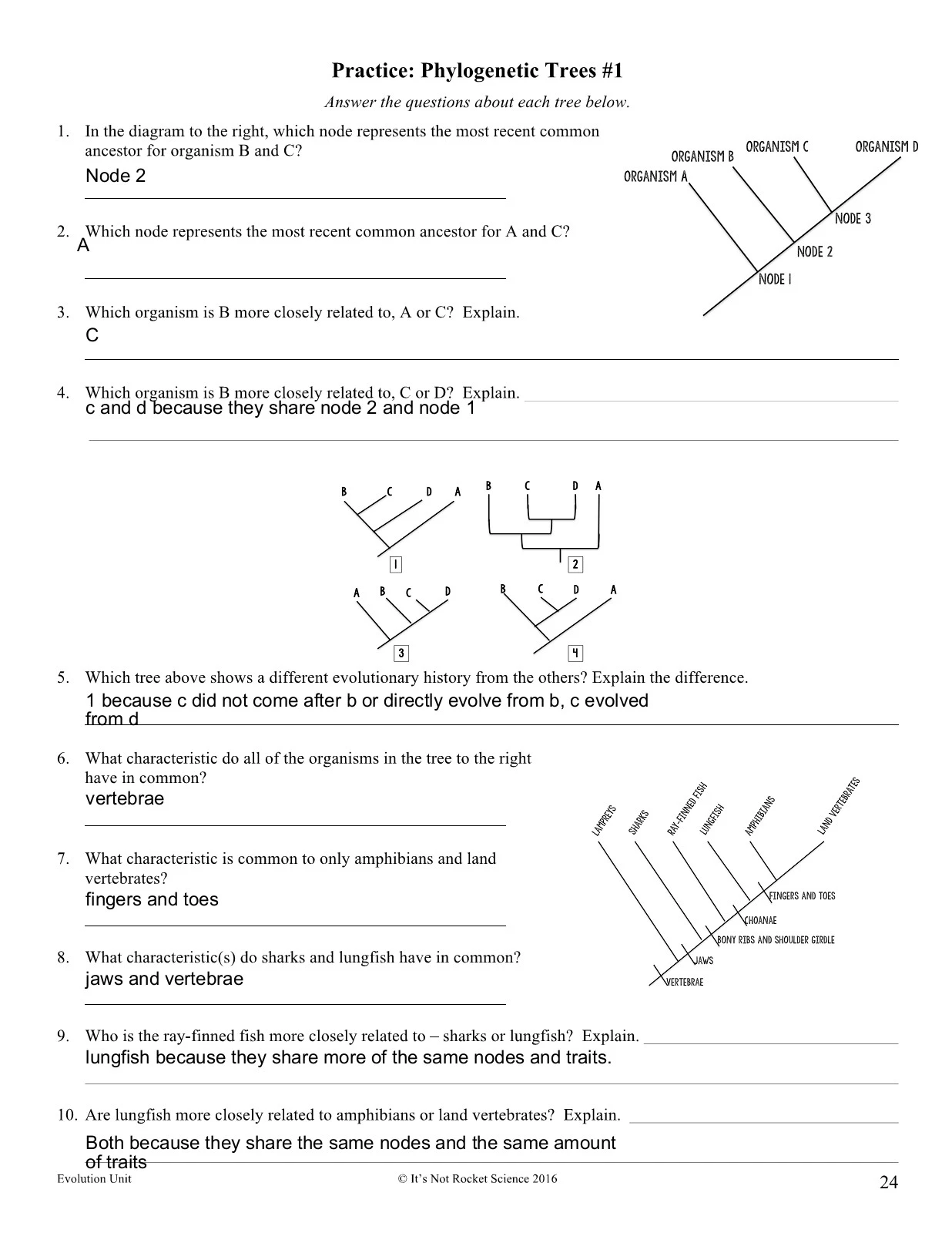
Creating a phylogenetic tree involves several steps:
- Select Data: Use genetic sequences, morphological traits, or fossil records to gather data for analysis.
- Alignment: Align sequences or characteristics to identify similarities and differences. For trees, this might mean leaf structure, bark texture, or DNA sequences.
- Model Selection: Choose a model of evolution that best describes how traits change over time. For example, DNA substitution models.
- Tree Construction: Apply algorithms like Maximum Parsimony, Maximum Likelihood, or Bayesian inference to construct the tree. Here’s how:
- Maximum Parsimony: Seeks the simplest explanation by minimizing evolutionary changes needed to explain the data.
- Maximum Likelihood: Considers different evolutionary pathways and selects the one most likely to have occurred.
- Bayesian: Incorporates prior probabilities to estimate the probability of different tree topologies.
- Interpretation: Once constructed, trees are interpreted to understand evolutionary relationships:
- Long branches suggest more evolutionary change.
- Short branches indicate closer relationships.
- Nodes help in deducing the timing of divergence.
Interpreting a Phylogenetic Tree

Phylogenetic trees, although simple in design, carry complex information. Here’s how you can interpret what they tell us:
- Clades: Groups of taxa that share a common ancestor are called clades. The monophyletic nature of clades helps in understanding classification and taxonomy.
- Branch Lengths: The length of the branches can indicate the amount of genetic divergence, the rate of evolution, or in some cases, time.
- Polytomy: A multi-pointed node in the tree, where more than two branches diverge, suggesting either a rapid divergence or incomplete resolution in the data.
Phylogenetic Tree of Trees Worksheet: Answer Key

Let’s dive into a sample worksheet designed to test understanding of phylogenetic trees with a focus on trees:
| Question | Answer |
|---|---|
| What does each node on the tree represent? | Each node represents a common ancestor from which descendent lineages diverged. |
| How does the length of branches indicate evolutionary relationships? | The branch length suggests the amount of change that has occurred since the species diverged from their common ancestor. |
| Explain the concept of a clade. | A clade is a group of organisms that includes an ancestor and all of its descendants. It represents a complete evolutionary lineage. |
| Why might two trees be very similar but have slightly different branch lengths? | This could result from different models of evolution used in tree construction or variations in data sources. |

🌱 Note: Remember that the actual construction of phylogenetic trees requires not just understanding but also software tools like MEGA, PAUP*, or MrBayes, which implement the algorithms discussed above.
In summarizing the journey through understanding and constructing phylogenetic trees:
Phylogenetic trees offer a window into the past, tracing the evolutionary paths species have taken. By learning how to construct and interpret these trees, especially for trees, we not only deepen our understanding of plant evolution but also contribute to fields like ecology, conservation biology, and even genetics. Trees, in this sense, provide insights into the evolutionary history of not just themselves but also their relationships with other species in the vast forest of life.
What do the terms ‘monophyletic’, ‘paraphyletic’, and ‘polyphyletic’ mean in the context of phylogenetic trees?
+
A monophyletic group includes an ancestor and all its descendants, representing a true lineage. Paraphyletic groups exclude some descendants, while polyphyletic groups consist of distantly related species with a common trait but not sharing a recent common ancestor.
Can phylogenetic trees be circular?
+
Yes, when represented in circular dendrograms or radial trees, phylogenetic relationships can be shown in a circular fashion, which can be useful for visualizing large datasets.
How does the choice of molecular marker affect the phylogenetic tree?

+
Different molecular markers evolve at different rates. For example, mitochondrial DNA changes faster than nuclear DNA, which can influence the resolution and interpretation of the evolutionary relationships depicted in the tree.
What is the significance of using outgroups in constructing phylogenetic trees?

+
An outgroup helps in rooting the tree by providing a point of comparison, allowing us to determine the direction of evolutionary change and better understand the relationships among the ingroup taxa.
How does horizontal gene transfer complicate the interpretation of phylogenetic trees?
+
Horizontal gene transfer introduces gene sequences from unrelated species into a lineage, creating challenges in accurately tracing evolutionary history, as trees might not reflect vertical descent alone.